While the article is being peer-reviewed, we have published the manuscript
in Pre-print format in the repository PeerJ Preprints, where it is freely
available at https://peerj.com/preprints/3429/
METABARCODING LITTORAL HARD-BOTTOM COMMUNITIES: UNEXPECTED DIVERSITY AND
DATABASE GAPS REVEALED BY TWO MOLECULAR MARKERS
Wangensteen OS, Palacín C,
Guardiola M, Turon X (2017) PeerJ Preprints 5:e3429v1
We report in this manuscript the astonishing diversity found in
communities from Islas Cies and Cabrera. We address several technical
aspects, compare the two genes studied (18S and COI), and perform
ecological analyses. Here goes the abstract:
We developed a metabarcoding method for biodiversity characterization of
structurally complex natural marine hard-bottom communities. Novel primer sets
for two different molecular markers: the “Leray fragment” of mitochondrial
cytochrome
c oxidase, COI, and the V7 region of ribosomal RNA 18S were
used to analyse eight different marine shallow benthic communities from two
National Parks in Spain (one in the Atlantic Ocean and another in the
Mediterranean Sea). Samples were sieved into three size fractions from where
DNA was extracted separately. Bayesian clustering was used for delimiting
molecular operational taxonomic units (MOTUs) and custom reference databases
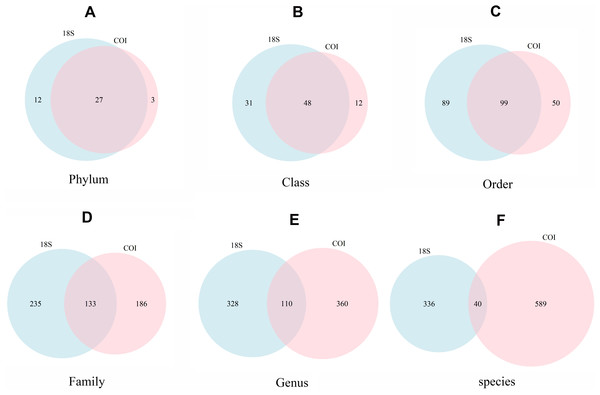
were constructed for taxonomic assignment. We found unexpectedly high values for
MOTU richness, suggesting that these communities host a large amount of yet
undescribed eukaryotic biodiversity. Significant gaps are still found in
sequence reference databases, which currently prevent the complete taxonomic
assignation of the detected sequences. Nevertheless, over 90% (in abundance) of
the sequenced reads could be successfully assigned to phylum or lower
taxonomical level. This identification rate might be significantly improved in
the future, as reference databases are updated. Our results show that marine
metabarcoding, currently applied mostly to plankton or sediments, can be
adapted to structurally complex hard bottom samples, and emerges as a robust,
fast, objective and affordable method for comprehensively characterizing the
diversity of marine benthic communities dominated by macroscopic seaweeds and
colonial or modular sessile metazoans, allowing for standardized biomonitoring
of these ecologically important communities. The new universal primers for COI
can potentially be used for biodiversity assessment with high taxonomic
resolution in a wide array of marine, terrestrial or freshwater eukaryotic
communities.
 |
Percent
of MOTUs found for the main categories of organisms in the three fractions
(A, B, C) studied with 18S (left) and COI (right) in all communities studied
|